Cloning:
4/13/14:
L-R reactions
Protocol:
- 1 μL of everything (μ = 03bc + alt + x in Word)
Conc:
Everything = 5 fmol (5 fmol per μL)
Destination vector = 10 fmol
- Put 1 μL of everything into the mix. Make sure all liquids coalesce
- Then add 1μL of Clonase
- After everything is added, pipet up and down. Careful not to add air bubbles. Then centrifuge. If air bubbles form: tap or manual centrifuge
- Incubate overnight at room temperature
Reaction tube numbers
- (Kyle) TRE-LacOid_mKate in DEST 1-2
- (Erik) Hef1a-lacO_ NLS-eYFP in DEST 1-2
- (Erik) Hef1a-lacO_ eBFP in DEST 1-2
- (Shinjini) CMV_mKATE in DEST 1-2
- (Shinjini) CMV_eYFP in DEST 1-2
4/15/14
Transformations
- Put competent cells from -80 degree Celsius freezer on ice
- Add 2μL of LR mixture to the cells. Do NOT pipet up and down; swirl instead.
- Keep cells on ice for 30 minutes.
- Heat shock cells at 42 degrees Celsius for EXACTLY 30 seconds
- Put it on ice for 2 minutes.
- The add .5mL (500μL) SOC
- Incubate at 37 degrees Celsius for 1 hour. (Tape cell tubes together and label!)
Making SOC
This is the ultra-broth for cell growth, so be extra careful not to get it contaminated.
- 49mL of SOB
- 1mL of 20% glucose.
Making Agar:
Follow instruction in pack.
Autoclave for around 20 minutes (+ ~20 minutes to pressurize and depressurize). Cool changing tape!
04/18/14
Update on LR's: LR's were left in the incubator for too long and they dried out. Trying to save 4 and 5.
PCR
Reaction Mixture:
- 1uL Template (entry vector eYFP_151ts; ie miRNA 451 target sites)
- Primers (comes in 100uM concentration) corresponding to the gene (2 primers)
- --> we need 2uL of 5uM conc. of each primer.
- --> we have to do 1–>20 dilution.
- 35uL of buffer/enzyme mix (comes in the pack)
Put reaction mixture into Thermocycler:
- Edit Settings
- initial Denature at 95 deg Celsius (for 5 mins)
- Cycles (35 cycles):
- 95 deg Celsius for 30sec
- 56 deg Celsius for 30 sec
- 72 deg Celsius for 30 sec
- Final annealing at 72 deg Celsius for 10 mins
- The hold at 4 deg Celsius indefinitely.
Labeling plates:
04/20/14
Making gel:
- TAE + Agarose powder
- 1% solution (1g/100mL)
- --> Using 50mL, so 0.5g of Agarose
- --> Added 0.496 - 0.506g
Microwave
- --> remember half-open cap!
- --> keep watch
- --> adjust power level (first higher, then lower)
- --> bring up to simmer till
- --> all melted
Once cool enough to touch (~10 mins)
- --> Add SYBR Safe (orange; liquid keep away from light)
- --> a dye that causes DNA to fluoresce; VERY important!
- --> turns it pink
- --> 10,000x working concentration (so, 5uL to 50mL)
Update on LR: plates didn't make it. Using backup plates
- 1-2_CMV_mKate
- 1-2_TRE-t_mKate
6.  1-2_TRE-t_tagBFP
1-2_TRE-t_tagBFP
Picking Colonies:
14mL Round bottomed tubes
LB medium (3mL)
Antibiotic (1000x conc.)
--> important or else other bacteria can grow, and the bacteria may lose their plasmids
--> 3uL to 3mL medium
- Take 2uL pipet and tip
- Scoop up colony (try not to get agar)
- Swirl and pipet up and down (Kyle)
- OR, leave pipet tip there (Katie)
Labeling Tubes:
"1-2_CMV_mKate" (1-2 is the destination vector, CMV is the promoter and mKate is the gene)
Running gel:
Check terminals (runs form +ve to -ve)
Fill with TAE till it covers the agarose gel (can leave the tray in)
Add dye (orange G)
--> 6x conc. so 8uL to 40uL (note, here the volume of dye is large enough to affect total conc.)
- Ladder
- 1 (15 uL) (for imaging) (1 = Left, EE)
- 2 (15 uL) (for imaging) (2 = Right, SS)
- Space
- 1 (~30 uL; fill as much as you can) (for extraction)
- 2 (~30 uL; fill as much as you can) (for extraction)
Wait till about halfway down ~30 mins
Extraction from gel:
- Image the gel
- Cut the extraction bands under UV (to see the bands)
- Follow protocol
04/20/14
Extraction from gel (similar to miniprep):
--> Weight
- 106mg
- 96mg
--> Add 3x vol of QG
- 318uL (1mg of agarose --> 1uL)
- 288uL
-->put in heat bath to melt (50 deg Celsius for ~10 mins; vortex every 2-3 mins?)
--> 1x vol of Isopropanol (original volume of gel)
- 106uL
- 96uL
-->Centrifuge at 13,000 rpm for 1 min; discard flowthrough
--> Add 500uL of QG
--> Centrifuge at 16,000 rpm for 1 min; discard flowthrough
--> Add 750uL of PE
--> Centrifuge again, I assume (not written in notes); dump PE
--> Dry run centrifuge
--> Add 50uL EB (elution buffer), let sit for 1 min (make sure to get it on the filter!)
--> Centrifuge; DO NOT discard flowthrough (this contains the DNA)
--> Nanodrop to measure concentration
Protip on pipets:
Try to work with 1 hand and not move the pipet much. Eg, take something, open with one hand, pipet things in, place it back down on the rack, etc.
05/16/14
Gel!
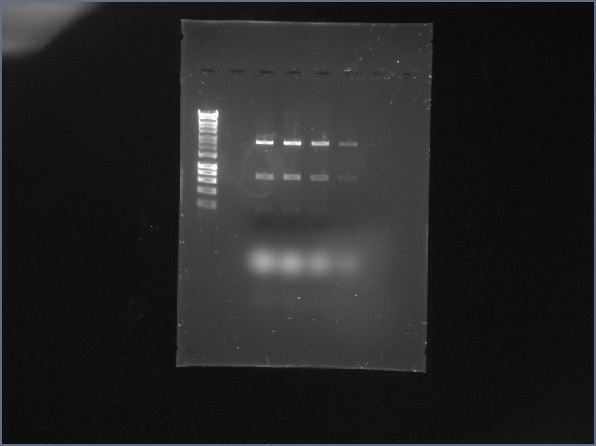
Summer Work:
9 June, Monday
Group Split
Wiki organization
10 June, Tuesday
More wiki planning
Planning initial experiments
11 June, Wednesday
Presented idea about using macrophages as vectors for the circuit
More planning and wiki stuff
Made logo
12 June, Thrusday
Golden Gated LilrB2, PirB and GGDonr
13 June, Friday
Transformed bacteria with LirB2/PirB/Digested GGDonr
14 June, Saturday
Put plate in fridge
15 June, Sunday
Picked colonies
Read on microglia
16 June, Monday
Cultures didn't grow
Read on microglia
17 June, Tuesday
Presented idea about potential use of microglia as treatment; Brian said he was more excited about using them as vectors
Cultures didn't grow
Troubleshooting:
- Transformed old GGDonr and New GGDonr (from Brandon) in old and new plates
18 June, Wednesday
Troubleshooting:
- Plates looked like this:
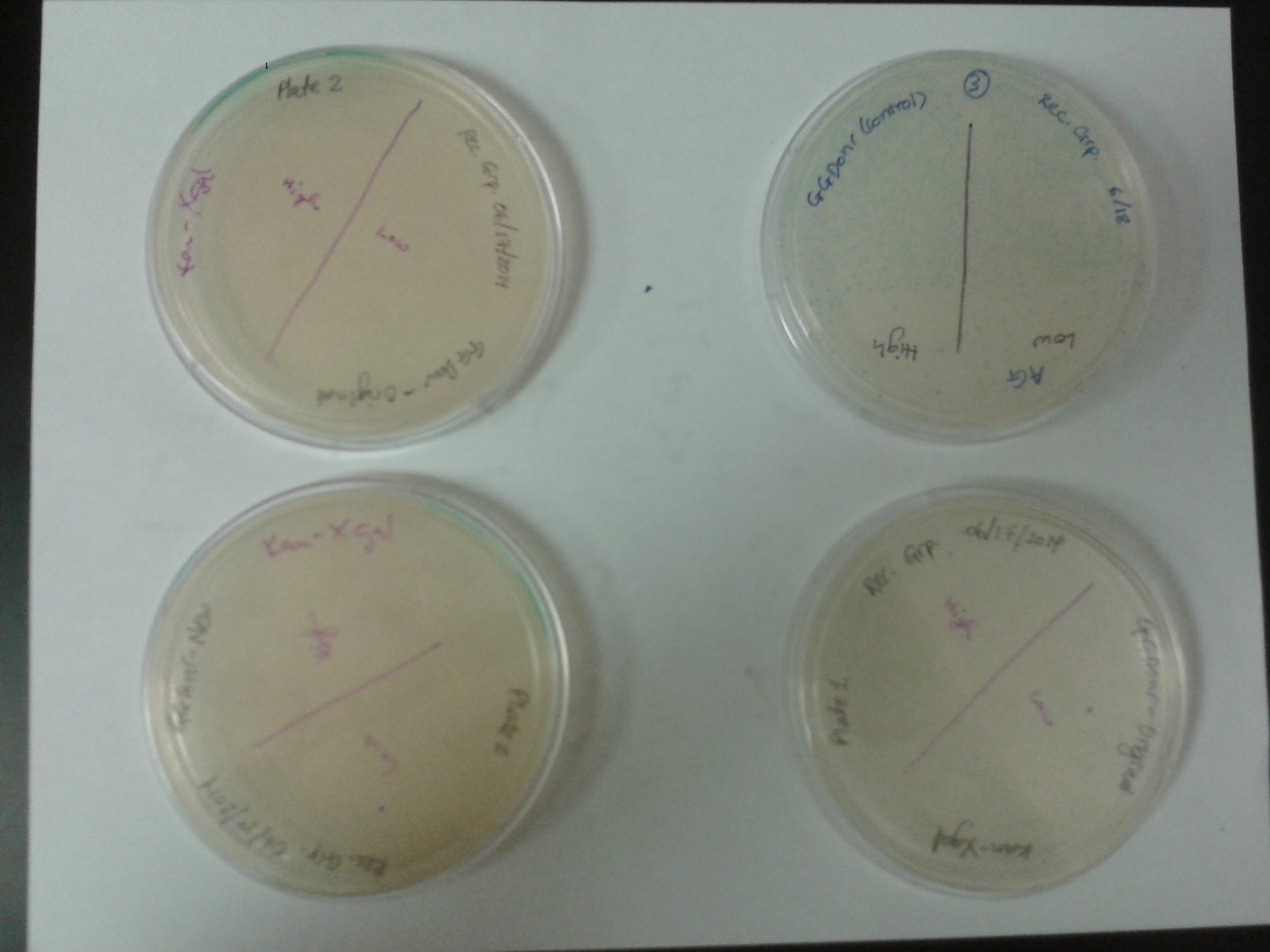
- No blue on old plates, blue on new plates, but much more colonies from new GGDonr. (New GGDonr on new plate was missing so I placed another plate that contained basically the same thing (new GGDonr replated as control).)
- Concultion: Old plates had some problems.
Transformed GG products into competent cells and plated again
Plated GGDonr as control
19 June, Thursday
PirB did not have white colonies
Picked white colony from LilrB2
Picked blue colony from GGDonr
Interacted with Mexico Highschool iGEM team; went to iGEM headquarters; showed kids around campus
GoldenGated LilrB2 and PirB again
20 June, Friday
Took out liquid cultures from incubator
NEGEM
22 June, Sunday
Miniprepped old LilrB2 and good GGDonr. Needs verification.
23 June, Monday
Showed Brian idea about silencing EP2 in microglia
Brian thinks interesting idea, but FIRST, need to look into
- Whether any engineering in microglia done
- whether there are microglia cell-lines
- protocols to grow them and stuff
because that's gonna take quite some time to get the cells and other culturing materials if they need special things, and basic fluorescence transfection, etc experiments need to be done on them.
It would be a good idea to find if anyone in the brain/cogsci labs is working on transforming microglia and "pick their brains".
Other things: verify miniprepped DNA
24 June, Tuesday
25 June, Wednesday
26 June, Thursday
27 June, Friday
28 June, Saturday
29 June, Sunday
Plated 3rd GG pENTR_LilrB2, pENTR_PirB.

